Sample a number of workers - used when nInd = NULL
(see SimParamBee$nWorkers).
This is just an example. You can provide your own functions that satisfy your needs!
nWorkersPoisson(colony, n = 1, average = 100)
nWorkersTruncPoisson(colony, n = 1, average = 100, lowerLimit = 0)
nWorkersColonyPhenotype(
colony,
queenTrait = 1,
workersTrait = NULL,
checkProduction = FALSE,
lowerLimit = 0,
...
)Arguments
- colony
- n
integer, number of samples
- average
numeric, average number of workers
- lowerLimit
numeric, returned numbers will be above this value
- queenTrait
numeric (column position) or character (column name), trait that represents queen's effect on the colony phenotype (defined in
SimParamBee- see examples); if0then this effect is 0- workersTrait
numeric (column position) or character (column name), trait that represents workers's effect on the colony phenotype (defined in
SimParamBee- see examples); if0then this effect is 0- checkProduction
logical, does the phenotype depend on the production status of colony; if yes and production is not
TRUE, the result is abovelowerLimit- ...
other arguments of
mapCasteToColonyPheno
Value
numeric, number of workers
Details
nWorkersPoisson samples from a Poisson distribution with a
given average, which can return a value 0. nDronesTruncPoisson
samples from a zero truncated Poisson distribution.
nWorkersColonyPhenotype returns a number (above lowerLimit)
as a function of colony phenotype, say queen's fecundity. Colony phenotype
is provided by mapCasteToColonyPheno. You need to set up
traits influencing the colony phenotype and their parameters (mean and
variances) via SimParamBee (see examples).
Functions
nWorkersTruncPoisson(): Sample a non-zero number of workersnWorkersColonyPhenotype(): Sample a non-zero number of workers based on colony phenotype, say queen's fecundity
See also
SimParamBee field nWorkers and
vignette(topic = "QuantitativeGenetics", package = "SIMplyBee")
Examples
nWorkersPoisson()
#> [1] 96
nWorkersPoisson()
#> [1] 89
n <- nWorkersPoisson(n = 1000)
hist(n, breaks = seq(from = min(n), to = max(n)), xlim = c(0, 200))
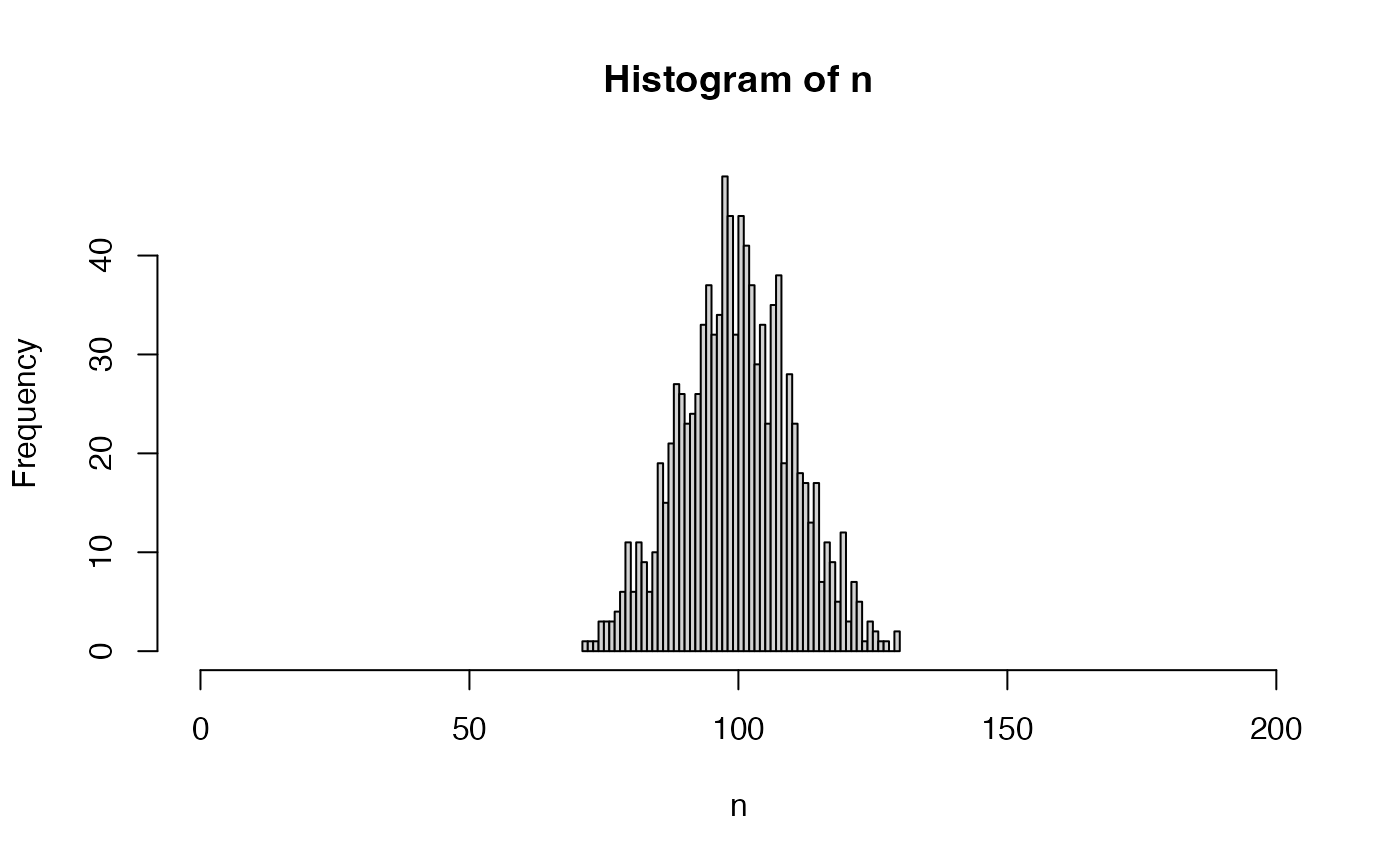 table(n)
#> n
#> 71 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91
#> 1 1 1 3 3 3 4 6 11 6 11 9 6 10 19 15 21 27 26 23
#> 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111
#> 24 26 33 37 32 34 48 44 32 44 41 37 29 33 23 35 38 19 28 23
#> 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 130
#> 18 17 13 17 7 11 9 5 12 3 7 5 1 3 2 1 1 2
nWorkersTruncPoisson()
#> [1] 117
nWorkersTruncPoisson()
#> [1] 112
n <- nWorkersTruncPoisson(n = 1000)
hist(n, breaks = seq(from = min(n), to = max(n)), xlim = c(0, 200))
table(n)
#> n
#> 71 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91
#> 1 1 1 3 3 3 4 6 11 6 11 9 6 10 19 15 21 27 26 23
#> 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111
#> 24 26 33 37 32 34 48 44 32 44 41 37 29 33 23 35 38 19 28 23
#> 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 130
#> 18 17 13 17 7 11 9 5 12 3 7 5 1 3 2 1 1 2
nWorkersTruncPoisson()
#> [1] 117
nWorkersTruncPoisson()
#> [1] 112
n <- nWorkersTruncPoisson(n = 1000)
hist(n, breaks = seq(from = min(n), to = max(n)), xlim = c(0, 200))
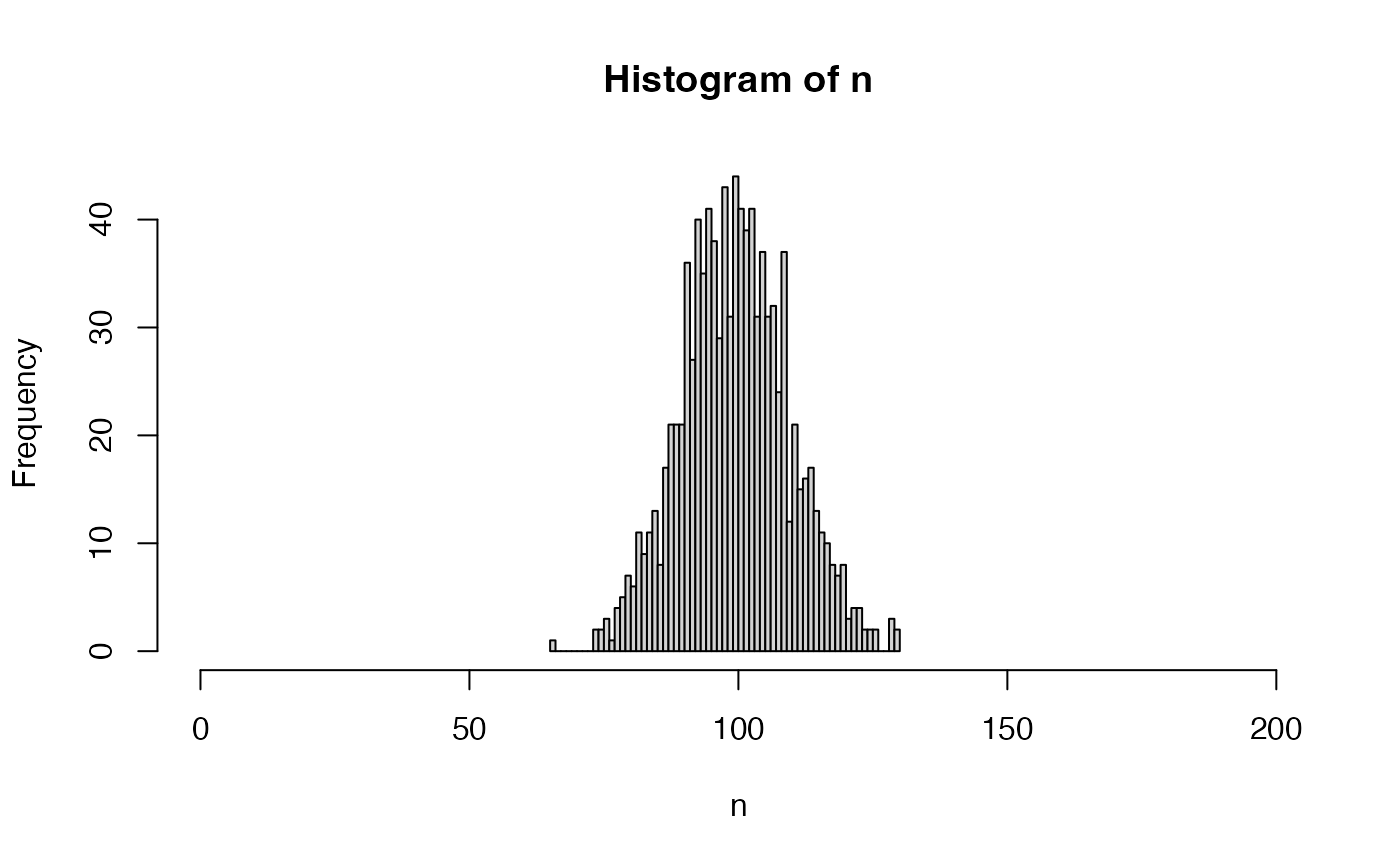 table(n)
#> n
#> 65 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92
#> 1 2 2 3 1 4 5 7 6 11 9 11 13 8 17 21 21 21 36 27
#> 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112
#> 40 35 41 38 29 43 31 44 41 39 41 31 37 31 32 24 37 12 21 15
#> 113 114 115 116 117 118 119 120 121 122 123 124 125 126 129 130
#> 16 17 13 11 10 8 7 8 3 4 4 2 2 2 3 2
# Example for nWorkersColonyPhenotype()
founderGenomes <- quickHaplo(nInd = 3, nChr = 1, segSites = 100)
SP <- SimParamBee$new(founderGenomes)
average <- 100
h2 <- 0.1
SP$addTraitA(nQtlPerChr = 100, mean = average, var = average * h2)
SP$setVarE(varE = average * (1 - h2))
basePop <- createVirginQueens(founderGenomes)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
drones <- createDrones(x = basePop[1], nInd = 50)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
droneGroups <- pullDroneGroupsFromDCA(drones, n = 2, nDrones = 15)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1 <- createColony(x = basePop[2])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony2 <- createColony(x = basePop[3])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1 <- cross(colony1, drones = droneGroups[[1]])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony2 <- cross(colony2, drones = droneGroups[[2]])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1@queen@pheno
#> Error in eval(expr, envir, enclos): object 'colony1' not found
colony2@queen@pheno
#> Error in eval(expr, envir, enclos): object 'colony2' not found
createWorkers(colony1, nInd = nWorkersColonyPhenotype)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
createWorkers(colony2, nInd = nWorkersColonyPhenotype)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
table(n)
#> n
#> 65 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92
#> 1 2 2 3 1 4 5 7 6 11 9 11 13 8 17 21 21 21 36 27
#> 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112
#> 40 35 41 38 29 43 31 44 41 39 41 31 37 31 32 24 37 12 21 15
#> 113 114 115 116 117 118 119 120 121 122 123 124 125 126 129 130
#> 16 17 13 11 10 8 7 8 3 4 4 2 2 2 3 2
# Example for nWorkersColonyPhenotype()
founderGenomes <- quickHaplo(nInd = 3, nChr = 1, segSites = 100)
SP <- SimParamBee$new(founderGenomes)
average <- 100
h2 <- 0.1
SP$addTraitA(nQtlPerChr = 100, mean = average, var = average * h2)
SP$setVarE(varE = average * (1 - h2))
basePop <- createVirginQueens(founderGenomes)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
drones <- createDrones(x = basePop[1], nInd = 50)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
droneGroups <- pullDroneGroupsFromDCA(drones, n = 2, nDrones = 15)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1 <- createColony(x = basePop[2])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony2 <- createColony(x = basePop[3])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1 <- cross(colony1, drones = droneGroups[[1]])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony2 <- cross(colony2, drones = droneGroups[[2]])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony1@queen@pheno
#> Error in eval(expr, envir, enclos): object 'colony1' not found
colony2@queen@pheno
#> Error in eval(expr, envir, enclos): object 'colony2' not found
createWorkers(colony1, nInd = nWorkersColonyPhenotype)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
createWorkers(colony2, nInd = nWorkersColonyPhenotype)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found